Out in Nature Nanotechnology, our work leverages a versatile molecular circuit with droplet microfluidics for the detection, quantification, and analysis of enzymes at the single molecule level [1].
The article can be accessed by following this link: https://rdcu.be/dzDGz
The idea of digital enzyme quantification, where single molecules are isolated in compartments to be detected individually, is not new and has been demonstrated by Boris Rotman in the ’60s with β-galactosidase [2].
Since then, this principle has been applied to a handful of fast catalysts (e.g. alkaline phosphatase or horseradish peroxidase) [3]. This is because such assays rely on the detection of the accumulation of – fluorescent – product, which is a linear process.
This work breaks this limit by coupling a target enzyme to an exponential and isothermal nucleic acid amplification process.
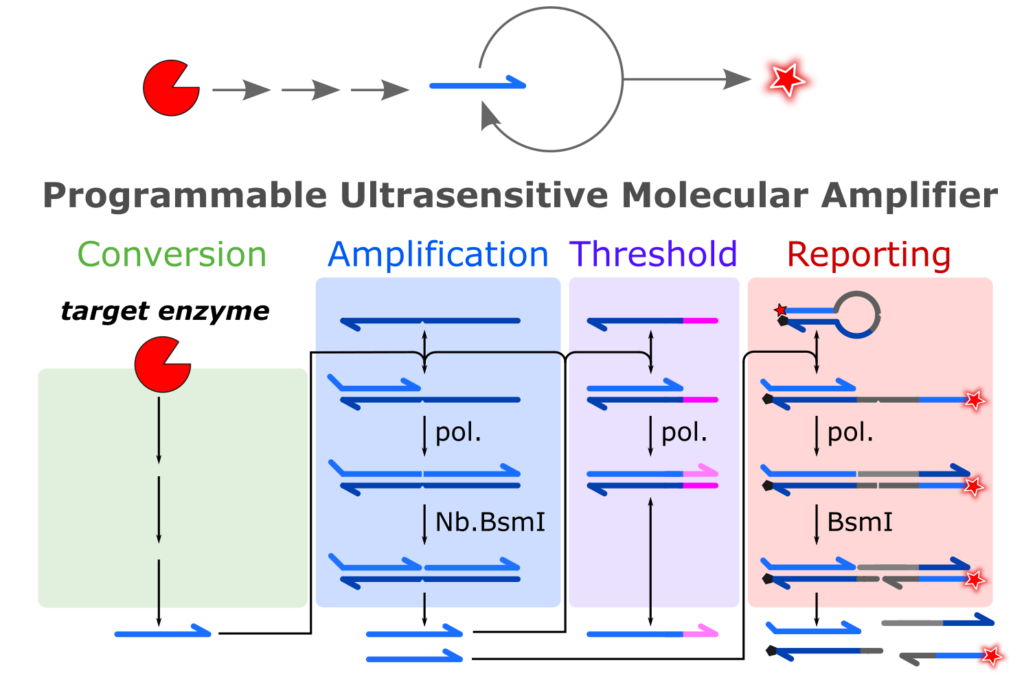
For all the enzymes we applied it to, the strategy is always the same. We designed an enzymatic cascade that links the target activity to the production of a short oligonucleotide. This oligo undergoes exponential replication by a molecular amplifier which leads to a strong fluorescence signal. We called these assay PUMA that stands for Programmable Ultrasensitive Molecular Amplifier.
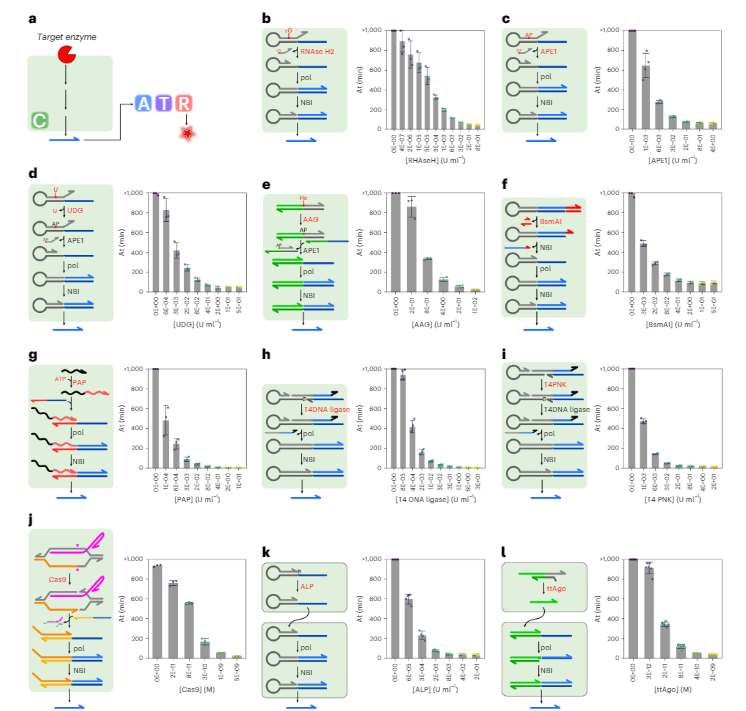
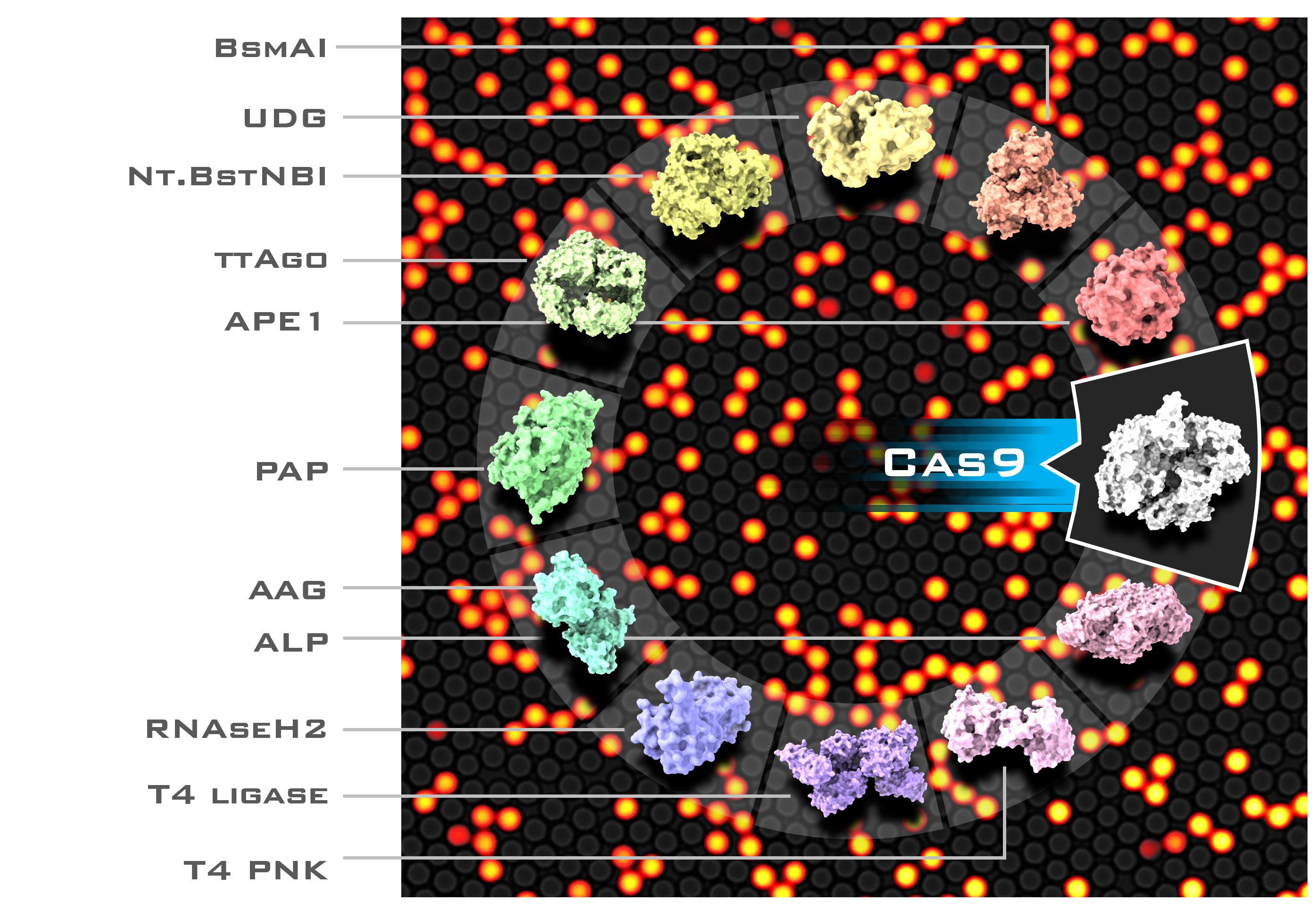
In principle, any enzyme compatible with the molecular circuit can be detected, irrespective of its turnover rate (even for suicide enzymes with single turnover !!). We design a dozen PUMA circuits for the sensitive detection of restriction enzymes, AP-endonucleases, RNAse, DNA glycosylase, phosphatase, polyadenine polymerase, kinase, Cas9, and Argonaute.
Because they benefit from the exponential amplification process that makes the success of nucleic acid-based diagnostic (e.g. qPCR), PUMA assays are orders of magnitude more sensitive than the ones based on linear product accumulation.
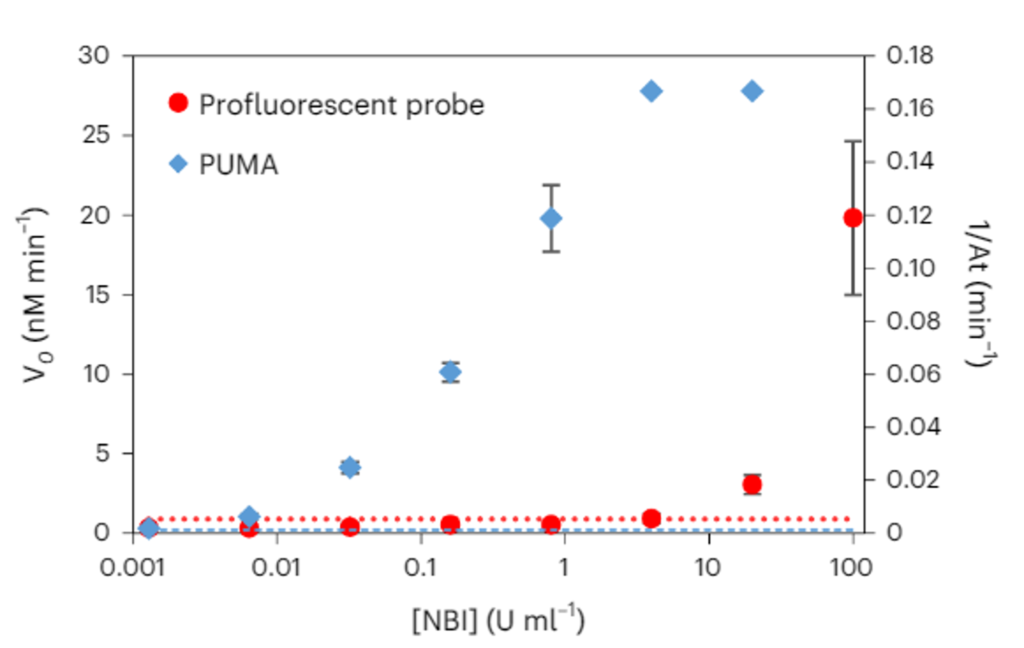
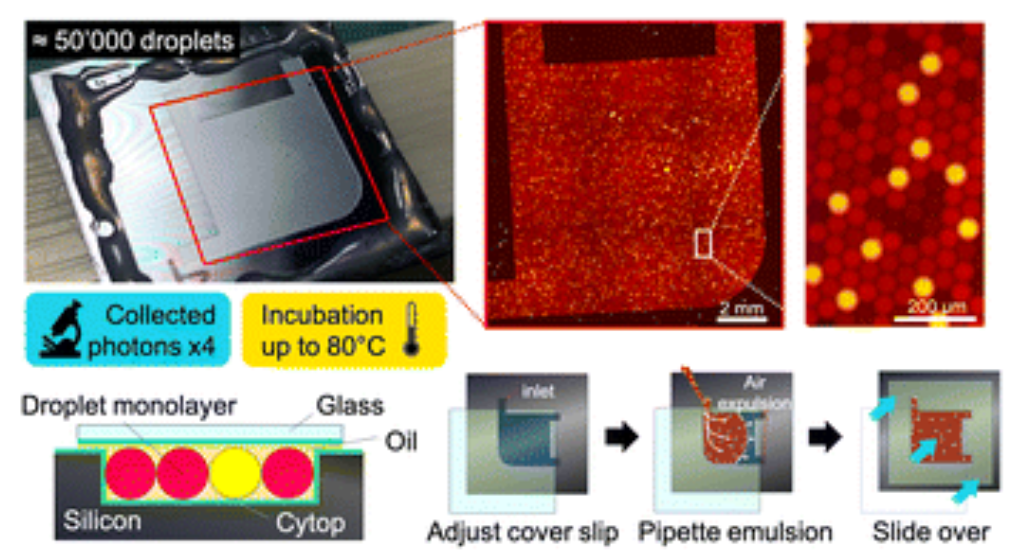
Another consequence of this gain of sensitivity, is that it gives us access to a digital format (dPUMA), where single enzymes are counted after being isolated in microfluidic droplets.
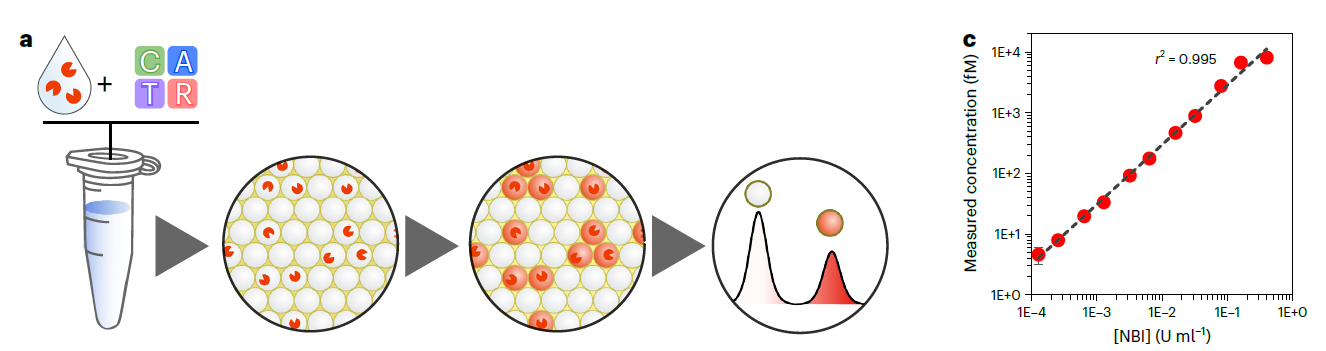
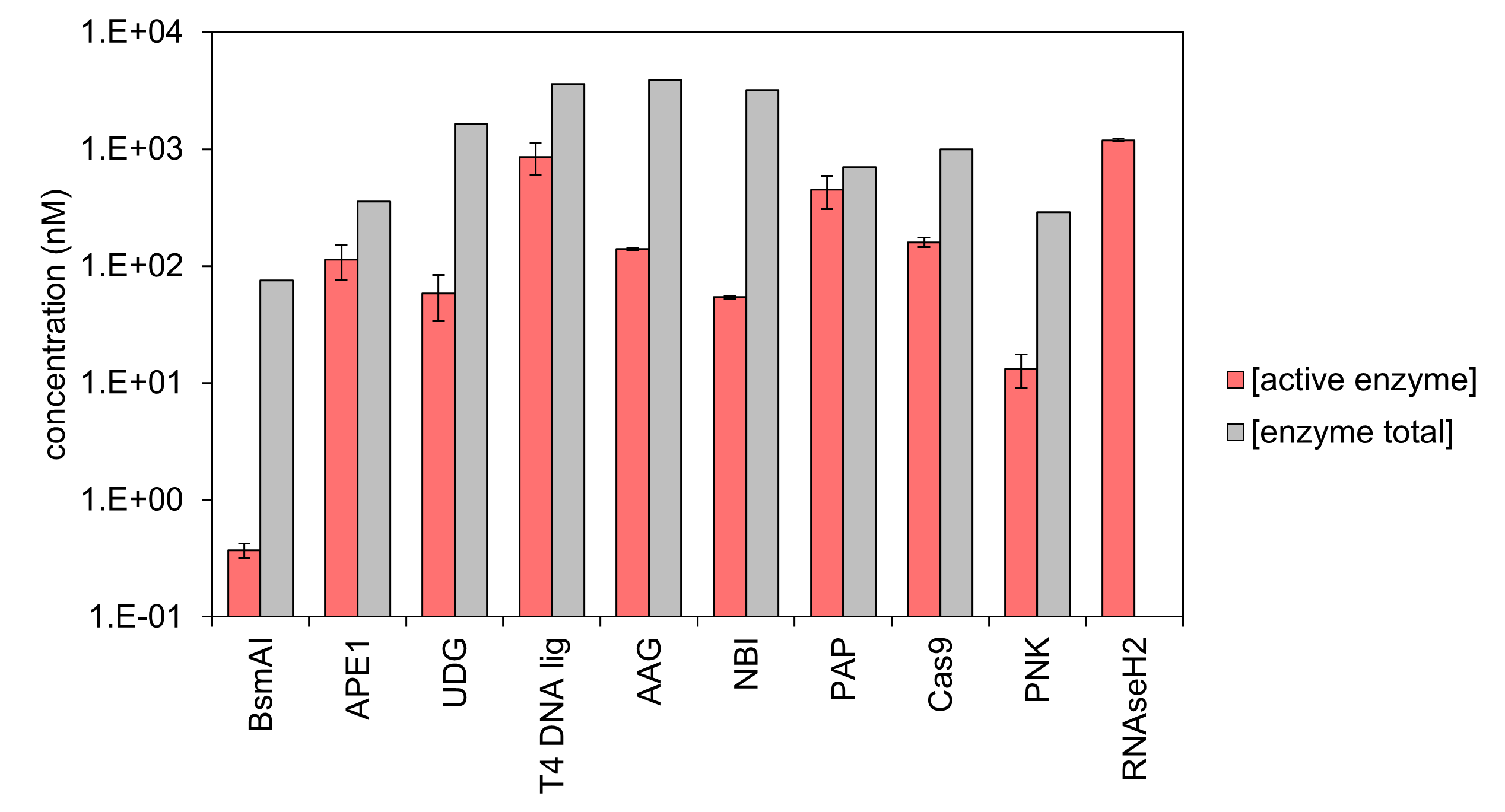
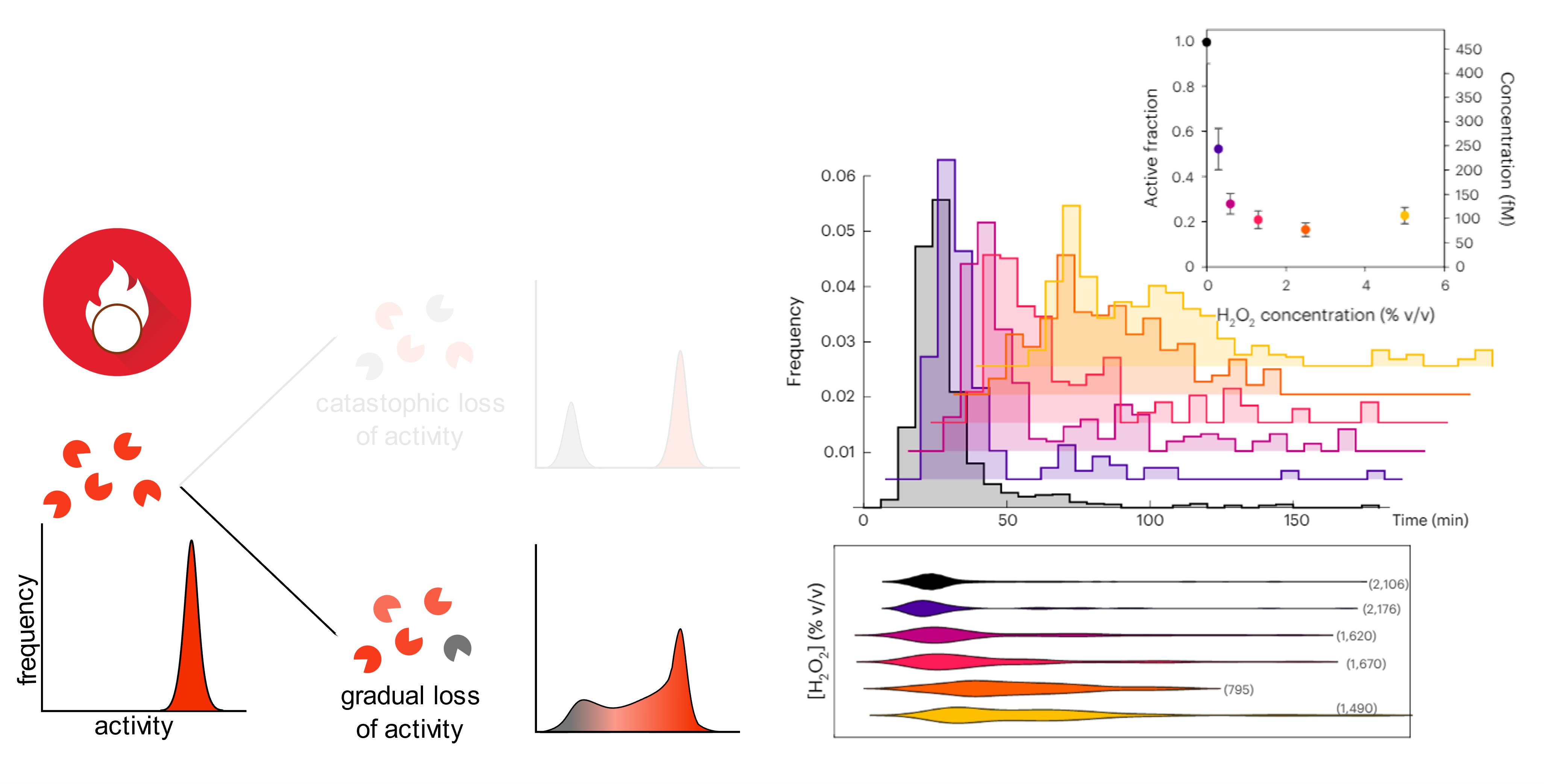
Interestingly, we observed that a large fraction of the purified enzyme is inactive by comparing the total concentration of enzymes (given by the provider) with the concentration of active enzyme measured with dPUMA.
Counting is good as it gives you the absolute concentration of active enzymes. But it does not tell you how active these enzymes are.
To access the activity distribution at the single-molecule level, we went one step further by recording in real-time the amplification reaction inside droplets. For each droplet that contains one enzyme, its amplification time is used as a proxy of the enzyme’s activity: the sooner it amplifies, the more active is the enzyme.
Special thanks to Nicolas Lobato-Dauzier et al. for developing silicon-based imaging chambers for precise temperature control [4].
We used this assay to look at the evolution of activity distribution of a homogeneous enzyme pool submitted to a thermal or oxidative stress.
Long story short: the thermal stress inactivates a substantial fraction of enzymes, but the remaining fraction is fully active (“catastrophic loss” mechanism) [5].
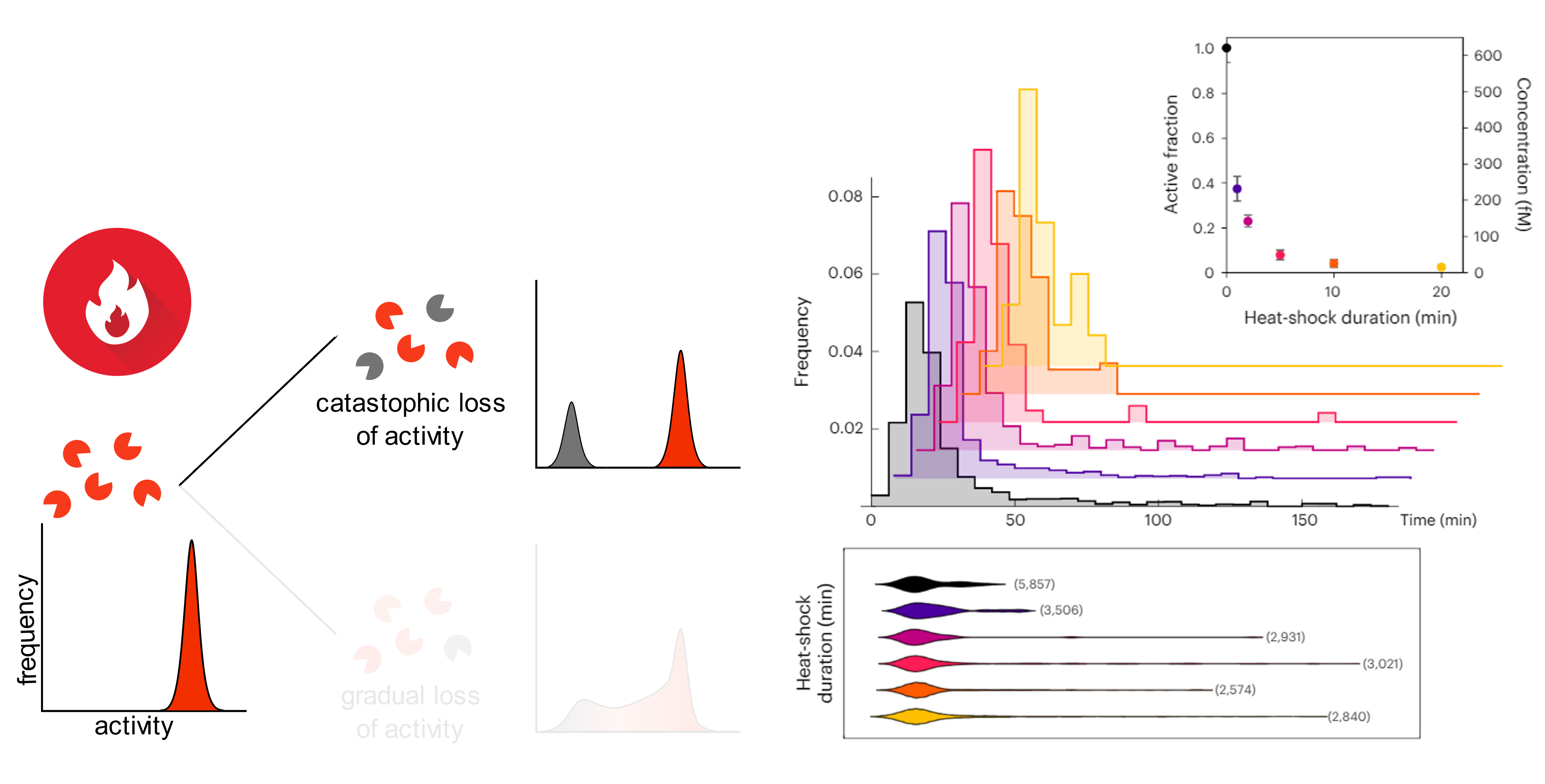
The oxidative stress (hydrogen peroxide treatment) also inactivates the enzyme, but the survivors exhibit a more spread activity distribution, with enzymes showing intermediate activity between fully active and dead (“gradual loss” of activity).
Another application is the fine functional characterization of mutants’ libraries, which may be of interest in the directed evolution of enzymes (see, the recent paper from our group [6]).
Take home message: dPUMA can count and analyze the functional diversity for a myriad of nucleic acid-related enzymes. It can be extended to many more provided a tailored conversion strategy.
We are now looking to apply such a technique for diagnostic (quantification of traces of enzymatic activity) or biotechnology (protein engineering).
To be continued…
This work is part of the objective one of the European Project MoP-MiP
References
[1] Gines, G. et al. Functional analysis of single enzymes combining programmable molecular circuits with droplet-based microfluidics, Nat. Nanotechnol. (2024) LINK
[2] Rotman, B. Measurement of activity of single molecules of β-d-galactosidase. Proc. Natl Acad. Sci. USA 47, 1981–1991 (1961).
[3] Noji, H., Minagawa, Y. & Ueno, H. Enzyme-based digital bioassay technology—key strategies and future perspectives. Lab Chip 22, 3092–3109 (2022).
[4] Lobato-Dauzier, N. et al. Silicon chambers for enhanced incubation and imaging of microfluidic droplets. Lab Chip 23, 2854–2865 (2023).
[5] D. B., Arriaga, E. A., Wong, J. C. Y., Lu, H. & Dovichi, N. J. Studies on Single alkaline phosphatase molecules: reaction rate and activation energy of a reaction catalyzed by a single molecule and the effect of thermal denaturation—the death of an enzyme. J. Am. Chem. Soc.18, 5245–5253 (1996).
[6] Dramé-Maigné, A. et al. In vitro enzyme self-selection using molecular programs. ACS Synth. Biol. (2024).
Laisser un commentaire