Open positions
Context
MicroRNAs are short endogenous RNA strands that finely tune gene expression through posttranscriptional regulation. Stably released in bodily fluids, these RNA strands are foreseen as promising liquid biopsies biomarkers in a wide range of clinical applications. Analytical challenges, however, remain to be addressed, including sensitivity, specificity, multiplexing, and quantification.
Our team at the Gulliver laboratory, located in the heart of Paris’ 5th district (ESPCI Paris, PSL Research University/CNRS) is looking for talented and motivated PhD students and postdocs to work on microRNA detection technologies. These positions are funded by an ERC starting grant (MoP-MiP). The project’s objective is to combine molecular programming concepts and microfluidics to build the next generation of microRNA detection assays. Join our dynamic team to work on DNA nanotechnology and enhance your skills in molecular biology and microfluidics!
For more background information, please visit: https://blog.espci.fr/guillaumegines/erc-mop-mip/
Publications or interest: https://blog.espci.fr/guillaumegines/publications/
Contact: Send CV and cover letter to guillaume.gines[at]espci.fr
Activities
- Conduct literature review and compile data.
- Plan and carry out biochemical experiments, including the assembly of biochemical reaction networks.
- Analyze data and images.
- Provide regular (weekly) progress reports.
- Engage in scientific communication (writing of scientific publications, conference attendance).
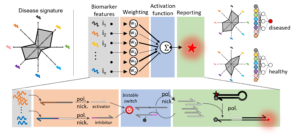
Position 1: DNA-enzyme neural networks for sample classification on microRNA concentrations
The candidate will focus on the design and experimental implementation of sophisticated biochemical circuits, akin in their architecture to neural networks, capable of integrating the concentration of many microRNAs and report a response according to the concentration pattern1. In collaboration with the LIMMS (University of Tokyo/CNRS), she/he will build on the strong expertise of the laboratory on the conception and assembly of molecular programs made out of DNA oligonucleotides and enzymes. The objectives of the research are i) to review the literature for defining robust microRNA classifiers for cancer monitoring, ii) to design and optimize in-moleculo microRNA classifiers and iii) validate the approach on clinical samples.
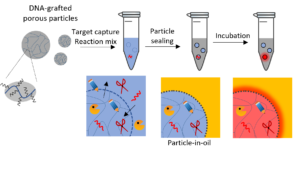
Position 2: Particle-templated emulsion for high throughput droplet digital bioassays
Droplet digital nucleic acid detection relies on the compartmentalization of individual targets into water-in-oil droplet prior to target amplification. Despite the multiple advantages (absolute quantification, high sensitivity, calibration-free), such assays rely on complex microfluidic setups that limit sample throughput and accessibility to non-expert users. Templated emulsions have been developed to palliate these limitations2,3. Here, hydrogel particles are used as pre-compartments that can be quickly sealed by mixing them vigorously with oil. Using monodisperse particles allows to create monodisperse, microfluidics-free droplets.
The candidate will develop and optimize this technology and adapt it to the original amplification method our team has developed for multiplex microRNA quantification.
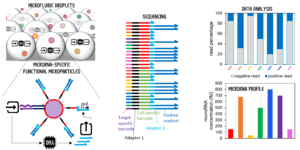
Position 3: Molecular programming and sequencing readout for accurate and multiplexed microRNA quantification.
The candidate will focus on an original methodology that performs digital and multiplex microRNA detection. The workflow implies microparticle functionalization with DNA strands, droplet microfluidics, and next-generation sequencing. The candidate will build on the solid expertise of the laboratory in the conception and assembly of molecular programs made of DNA oligonucleotides and enzymes, capable of information processing.
References
- Okumura, S. et al. Nonlinear decision-making with enzymatic neural networks. Nature 610, 496–501 (2022).
- Hatori, M. N., Kim, S. C. & Abate, A. R. Particle-Templated Emulsification for Microfluidics-Free Digital Biology. Anal. Chem. 90, 9813–9820 (2018).
- de Rutte, J. et al. Suspendable Hydrogel Nanovials for Massively Parallel Single-Cell Functional Analysis and Sorting. ACS Nano (2022) doi:10.1021/acsnano.1c11420.
