credit cover art: Thomas Maniaque
Better latter than never ! We reported earlier this year a new technology termed Digiplex designed to accurately measure multiple microRNAs at the single-molecule level.
Available on JACS : https://pubs.acs.org/doi/abs/10.1021/jacs.5c07214
Modern medicine increasingly relies on molecular markers (mainly biomolecules found in patient samples) to detect diseases earlier and guide treatment. These biomarkers can include genetic mutations, epigenetic modifications, or changes in the levels of specific proteins or RNA molecules.
Among these RNA molecules are microRNAs: short pieces of RNA that do not code for proteins but play a crucial role in regulating gene expression. Dysregulated microRNAs have been linked to a wide range of diseases, from cancer to neurodegenerative disorders. Measuring many microRNAs at once, referred as multiplexing, can provide a detailed picture of what’s happening inside a cell or tissue, offering helpful information for diagnosis and patient monitoring.
The challenge: sensitivity, accuracy, and multiplexing
Despite their potential, microRNA tests are still rarely used in clinical settings. One major reason is the lack of a method that is both highly sensitive and able to measure many microRNAs simultaneously. Put simply, current technologies sit at two extremes. Next-generation sequencing (NGS) offers virtually unlimited multiplexing but comes with limited sensitivity and accuracy. At the other end of the spectrum are amplification-based methods such as RT-PCR (the same technique used for COVID testing). These are extremely sensitive but hard to multiplex (especially when dealing with very short RNA molecules like microRNAs). Accuracy can be further increased using a digital format, where the sample is mixed with amplification reagents and then partitioned into thousands of tiny droplets. Because molecules randomly distribute across droplets (according to what’s known as the Poisson distribution), each droplet essentially contains either zero or one target molecule. Yet despite this precision, digital PCR still struggles to measure many microRNAs simultaneously.
Our solution: Digiplex, a digital and multiplexed microRNA profiling method
In our recent study, we present Digiplex, a new technology designed to accurately measure multiple microRNAs at the single-molecule level. It combines three key components: Microbeads for multiplexing, an isothermal amplification chemistry (previously reported), and droplet microfluidics for isolating individual molecules.
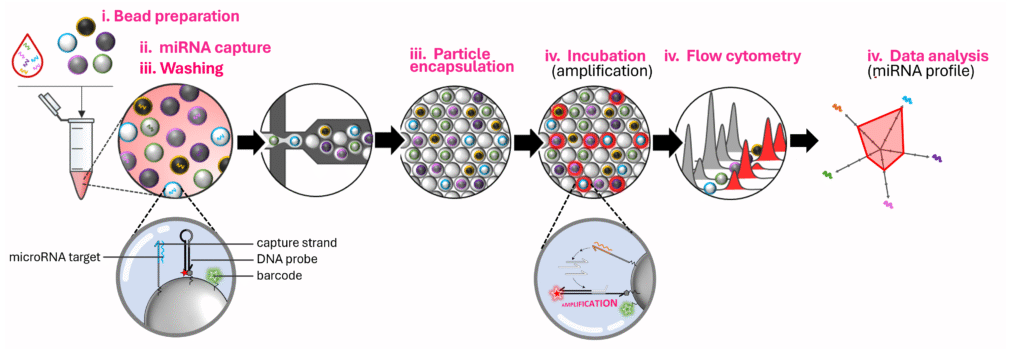
Here’s how the process works:
- Bead preparation: we create different groups of microbeads. Each group carries DNA strands designed to capture one specific microRNA, along with a unique fluorescent “barcode” that identifies the target, and a DNA probe for reporting the amplified signal.
- MicroRNA capture: all bead groups are mixed with the sample. MicroRNAs bind to their matching beads. Because of how molecules randomly distribute, each bead ends up with essentially either zero or one microRNA.
- Washing: we rinse away the rest of the sample and add our isothermal amplification mixture.
- Encapsulation: using a microfluidic chip, each bead is isolated inside a tiny droplet.
- Incubation: if a bead carries a microRNA, that single molecule triggers signal amplification, switching the bead “ON.” If not, it remains “OFF.”
- Flow cytometry: after recovering the beads from the droplets, these are analyzed one by one through a flow cytometer. The machine reads their fluorescent barcode (which microRNA they measure) and whether they are ON or OFF (whether the target was present)
- Data analysis: By counting the number of ON and OFF beads for each microRNA type, we can calculate its exact concentration and reconstruct the sample’s microRNA profile.
After carefully optimizing the method on a single target, we demonstrated a 10-plex assay, successfully profiling tissue extracts and comparing our results with digital PCR. Digiplex showed high accuracy while enabling greater multiplexing.
What comes next ?
Detecting 10 microRNAs at once is good, 100 would be better. We are working to increase the multiplexing capacity, robustness, and user-friendliness of Digiplex, with the long-term aim of making it a routine tool for both microRNA research and clinical use.
A big congratulations to our two first co-authors, Thomas Jet and Coline Kieffer, who relentlessly worked to solve the problems we faced over the last 8 years.
This work was supported by the European Research Council (ERC Starting Grant 949493), PSL Research University, INSERM, and the Région Île-de-France.
link to the article: https://pubs.acs.org/doi/abs/10.1021/jacs.5c07214

Laisser un commentaire