How to Use FirstGlance |
 Clicking on an atom produces an identification
report in the Jmol rectangle, to the lower left of the molecule (see
snapshot at right), and also in the browser's status bar.
(In
Firefox on Mac OSX only, you often have to click twice
before the report is produced.)
Amino acids are reported as
3-letter abbreviations,
and nucleotides as A, C, G, T, or U.
Ligands and non-standard residues have other abbreviations.
If you don't recognize the abbreviation, click on the PDB icon
(upper left panel in FirstGlance), and there,
the Biology & Chemistry tab, and look under
Ligands and Prosthetic Groups; or click the OCA
icon (upper left panel in FirstGlance), and
look under ligand.
Clicking on an atom produces an identification
report in the Jmol rectangle, to the lower left of the molecule (see
snapshot at right), and also in the browser's status bar.
(In
Firefox on Mac OSX only, you often have to click twice
before the report is produced.)
Amino acids are reported as
3-letter abbreviations,
and nucleotides as A, C, G, T, or U.
Ligands and non-standard residues have other abbreviations.
If you don't recognize the abbreviation, click on the PDB icon
(upper left panel in FirstGlance), and there,
the Biology & Chemistry tab, and look under
Ligands and Prosthetic Groups; or click the OCA
icon (upper left panel in FirstGlance), and
look under ligand.
|
CA = Carbon, Alpha CB = Carbon, Beta CG, OG = Carbon/Oxygen, Gamma CD = Carbon, Delta NE, OE = Nitrogen/Oxygen, Epsilon NZ = Nitrogen, Zeta NH = Nitrogen, Eta |
O3*, O5* = 3' and 5' pentose oxygens C1* to C5* = 1' to 5' pentose carbons C2-C8, N1-N9, O2-O6 = elements in bases C5M = C5 Methyl (in Thymine) |
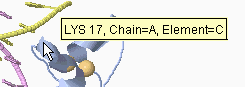 In addition to clicking, you can simply touch an atom with the mouse
(without moving the mouse, which is called "hovering"), and after a
delay of about two seconds, a yellow message will appear near the
mouse pointer (see snapshot at right) with an atom identification
report. This works best with Spin off.
In addition to clicking, you can simply touch an atom with the mouse
(without moving the mouse, which is called "hovering"), and after a
delay of about two seconds, a yellow message will appear near the
mouse pointer (see snapshot at right) with an atom identification
report. This works best with Spin off.
|
Ala A Alanine
Arg R Arginine Asn N Asparagine Asp D Aspartic acid (mnemonic: asparDic) Cys C Cysteine |
Gln Q Glutamine (mnemonic: Quetamine) Glu E Glutamic acid (mnemonic: gluEtamic) Gly G Glycine His H Histidine Ile I Isoleucine |
Leu L Leucine
Lys K Lysine (mnemonic: liKesine) Met M Methionine Phe F Phenylalanine (mnemonic: Fenylalanine) Pro P Proline |
Ser S Serine
Thr T Threonine Trp W Tryptophan (mnemonic: tWyptophan) Tyr Y Tyrosine Val V Valine |
***** SAMPLE FOR 2ACE *****
S C S C
SEQCRD 0 D ASP --- 1 - - - 367 Key to 3-Letter Codes
SEQCRD 0 D ASP --- 2 - - - 367
SEQCRD 0 H HIS --- 3 - - - 367 THIS LISTING IS NOT FOR YOUR MOLECULE!
SEQCRD 0 S SER SER 4 4 C T 5 EXAMPLE AT LEFT IS FOR 2ACE.
SEQCRD 0 E GLU GLU 5 5 C T 5 FOR YOUR MOLECULE,
[lines 6-482 omitted] SEQCRD 0 N ASN ASN 483 483 C T 5
SEQCRD 0 E GLU GLU 484 484 C T 5
SEQCRD 0 P PRO --- 485 - - - 367
SEQCRD 0 H HIS --- 486 - - - 367
SEQCRD 0 S SER --- 487 - - - 367
SEQCRD 0 Q GLN --- 488 - - - 367
SEQCRD 0 E GLU --- 489 - - - 367
SEQCRD 0 S SER SER 490 490 C T 5
SEQCRD 0 K LYS LYS 491 491 C T 5
[lines 492-533 omitted]
SEQCRD 0 A ALA ALA 534 534 H H -
SEQCRD 0 T THR THR 535 535 H C 5
SEQCRD 0 A ALA --- 536 - - - 367
SEQCRD 0 C CYS --- 537 - - - 367
The conclusion from the above listing is that residues
1-3, 485-489, and 536-537 lack coordinates.
In a long S2C listing such as the one above, you can use your browser's
Find in page with the search word "---" to jump to any gaps.
4, 484, 490, 535into the Find.. slot and press Enter. You will be able to see the gap in the chain, marked by halos. By clicking on the beginning and end of the chain (marked with halos) you will see that the chain does not start at 1, nor does it end at 537. Notice that without examining the S2C listing, it would have been easy to overlook the missing residues in the crystallographic model.

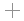


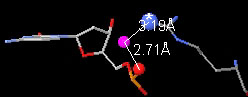 Water bridge in 1D66 between the sidechain nitrogen of Arg 46
(chain B)
and the phosphate in Cytosine 13 (chain D).
Water bridge in 1D66 between the sidechain nitrogen of Arg 46
(chain B)
and the phosphate in Cytosine 13 (chain D).
|
 Problem. FirstGlance will often fail to display
anomalous atoms appropriately. Some anomalous atoms will be invisible
in all views except Vines (with details on).
Problem. FirstGlance will often fail to display
anomalous atoms appropriately. Some anomalous atoms will be invisible
in all views except Vines (with details on).
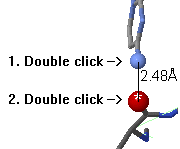 To measure the distance between two atoms, double click on the
first atom. Now, as you touch other atoms, the distances will be shown
on a red line connecting the atoms (except in Mac Firefox).
Double click on a second atom to fix the distance report.
To measure the distance between two atoms, double click on the
first atom. Now, as you touch other atoms, the distances will be shown
on a red line connecting the atoms (except in Mac Firefox).
Double click on a second atom to fix the distance report.
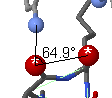 To measure an angle, double click on the first atom, then single click
on the second. As you touch additional atoms, the angle will be displayed
on a red line. Double click the third atom to fix the angle report.
To measure an angle, double click on the first atom, then single click
on the second. As you touch additional atoms, the angle will be displayed
on a red line. Double click the third atom to fix the angle report.
 To measure a torsion (dihedral) angle, follow the same kind of procedure,
except four atoms are needed.
To measure a torsion (dihedral) angle, follow the same kind of procedure,
except four atoms are needed.
<form method='post' enctype='multipart/form-data'
action='http://bip.weizmann.ac.il/oca-bin/fgij'>
Upload a PDB file of your own:
<br><small>
Caution: uploading a PDB file may <b>disclose your data</b> to others.
</small><br>
<input type='file' name='file' size='50'>
<input type='submit' value='View in FirstGlance'>
</form>